Kleiner develops method to profile RNA-modifying enzymes

Using a largescale approach to studying cellular proteins called chemoproteomics, Ralph Kleiner, assistant professor in the Department of Chemistry, has developed a methodology that provides insights into a cell’s RNA modifications and their associated enzymes, expanding our knowledge of the workings of this central biomolecule.
Enzymes are proteins that catalyze certain chemical reactions in cells. The modifications they install are perhaps of the greatest diversity in RNA. These modifications have different biological functions, but their significance is not fully understood.
The Kleiner lab’s research takes up that mission. The lab’s novel method is based on a chemical probe – actually an artificial molecule – that is essentially imbibed by cells as they manufacture RNA, allowing associated enzymes to be isolated.
“There’s kind of this mystery surrounding why biology is putting in all of these modifications onto RNA, this absolutely central molecule in biology. That’s not a new theme – there are sets of modifications that are put onto DNA and protein and those have been studied. But what people are realizing now is that the modifications on RNA are just as important,” said Kleiner.
“We wanted to develop a way to discover and characterize new RNA-modifying enzymes to gain insight into what these enzymes and associated modifications are doing.”
The method works so well, in fact, that researchers used it in the same study to isolate an enzyme called DUS3L, which installs the modification dihydrouridine on human RNA. Dihydrouridine has been known since the 1960s and is present in most organisms, but the human enzymes that install dihydrouridine have been poorly studied. Using their new method, researchers profiled RNA substrates of DUS3L for the first time.
The paper, “Activity-based RNA modifying enzyme probing reveals DUS3L-mediated dihydrouridylation,” was published this week in Nature Chemical Biology. The research is a first step in a proposal for which Kleiner was awarded a CAREER grant in 2019 by the National Science Foundation (NSF), Division of Molecular and Cellular Bioscience.
RNABPP
The chemical probing strategy developed by Kleiner is called RNA-mediated Activity-Based Protein Profiling (RNABPP). It relies on metabolic labeling and quantitative proteomics – enhanced with the tools of organic chemistry – to investigate RNA-modifying enzymes.
As with DNA, RNA is composed of four canonical letters. However, scientists now know that these letters can be chemically modified to form different letters that deepen RNA’s informational complexity and its role in cells. In fact, more than 150 RNA modifications have already been identified in organisms.
In Kleiner’s study, the chemical probe is essentially “fed” to living cells. Exploiting cellular metabolism, the interloper then makes its way into the chemical alphabet of RNA as a kind of building block, plucked up by cells as they manufacture RNA. This infiltration allows researchers to crosslink RNA with the specific enzymes that seek to modify it.
Varying the chemical structure of the probe affects the types of RNA-modifying enzymes that can be captured. The particular probe used in the current study allowed crosslinking of DUS3L as well as enzymes that add a methyl group to RNA.
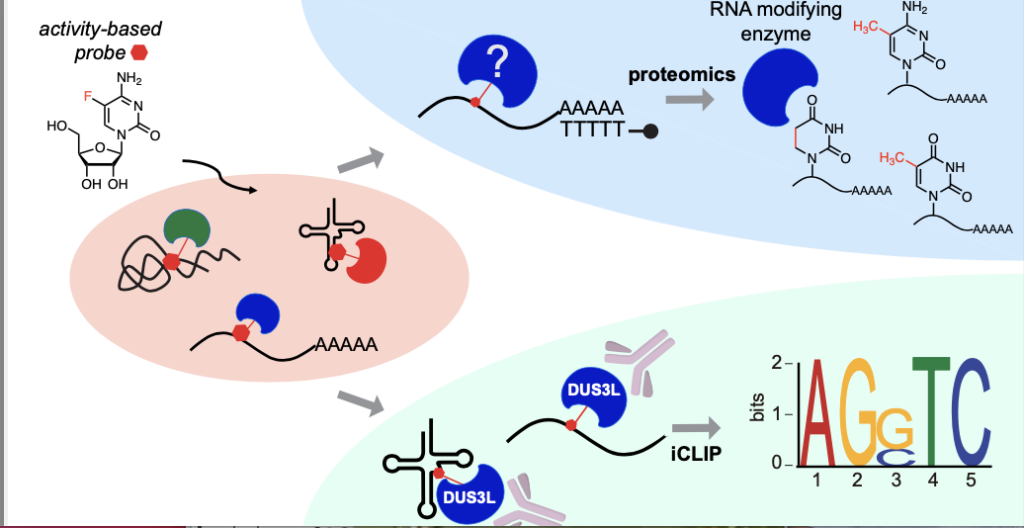
By incorporating an activity-based probe into cellular RNA, RNABPP allows the capture of RNA modifying enzymes in action. Combining with proteomics or RNA sequencing, the enzymes or modified transcripts can be identified
“An RNA-modifying enzyme is not going to randomly modify everything it comes across. Instead, it is going to choose – am I going to install this modification on tRNA or mRNA, and then which mRNA?” said Wei (Crystal) Dai, co-lead author on the paper and a graduate student in the Kleiner lab. “Our method uses the chemical probe that is going to react specifically with these types of enzymes. They then form a very stable crosslink with these enzymes, so now we can capture them – lock the enzymes with its modifying target and then basically pull out the RNA with attached protein.”
Subsequently, the Martin Wühr lab characterized the enzymes through proteomics, a mass spectroscopy-based method of looking at many thousands of cellular proteins all at once, in which they have extensive experience.
“I have been very impressed by the Kleiner Lab’s ability to leverage their chemical biology expertise to design novel experiments with proteomics and transcriptomics readouts,” said Martin Wühr, assistant professor in the Department of Molecular Biology and the Lewis-Sigler Institute for Integrative Genomics. “Integrating all this information, they were able to shine light on the dark corners of the RNA-modifying proteome.”
THE DUS3L IN THE HAYSTACK
Uridine, a pyrimidine nucleoside abbreviated simply as “U,” is one of the four canonical letters in RNA. Dihydrouridine is a modification of uridine. Dihydrouridine synthase 3L (DUS3L) is an enzyme that generates this modification. That synthase is what the Kleiner lab has discovered in this study through the RNABPP technique. It provides a compelling example of how the technique can identify and characterize enzymes and their substrates on the road to understanding their roles.
“The modification has been known for a very long time, but we still don’t know what it’s doing and how it’s regulated,” said Dai. “Therefore, we’ll be able to further explore its biological roles and functions.”
She added that future research collaborations will seek to further investigate the biological function of DUS3L.
“RNABPP is a very general method, and through the discovery of DUS3L, we’ve validated it,” said Kleiner. “We know that it works. We envision that we could apply this method to any kind of biological system to study its particular complement of RNA-modifying enzymes.”
Read the full Nature Chemical Biology paper.
Authors on this paper include leads Wei Dai and Ang Li; Nathan Yu, Thao Nguyen, Robert Leach, Martin Wühr, and Ralph Kleiner. Funding for this research was provided by a National Science Foundation CAREER award (MCB-1942565); the National Institute of Health (R01 GM132189); the Sidney Kimmel Foundation; and the Alfred P. Sloan Foundation. This work was also supported by NIH grant R35 GM128813 (to M.W.).
