Unlocking the Potential of Bacterial Gene Clusters to Discover New Antibiotics
Resistance to antibiotics has been steadily rising and poses a serious threat to the stronghold of existing treatments. Now, a method from Mohammad Seyedsayamdost, an assistant professor of chemistry at Princeton University, may open the door to the discovery of a host of potential drug candidates.
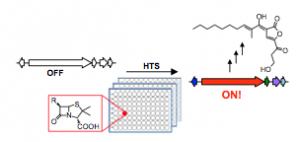
The vast majority of anti-infectives on the market today are bacterial natural products, made by biosynthetic gene clusters. Genome sequencing of bacteria has revealed that these active gene clusters are outnumbered approximately ten times by so-called silent gene clusters.
“Turning these clusters on would really expand our available chemical space to search for new antibiotic or otherwise therapeutically useful molecules,” Seyedsayamdost said.
In an article published last week in the journal Proceedings of the National Academy of Sciences, Seyedsayamdost reported a strategy to quickly screen whole libraries of compounds to find elicitors, small molecules that can turn on a specific gene cluster. He used a genetic reporter that fluoresces or generates a color when the gene cluster is activated to easily identify positive hits. Using this method, two silent gene clusters were successfully activated and a new metabolite was discovered.
Application of this work promises to uncover new bacterial natural products and provide insights into the regulatory networks that control silent gene clusters.
Read the full article here:
“High-throughput platform for the discovery of elicitors of silent bacterial gene clusters.” Seyedsayamdost, M. R. Proc. Natl. Acad. Sci. 2014, Early edition.
