Mo Lab: New Pathway for Antimicrobial Peptides
Researchers in the Mo Lab have discovered a new multi-step pathway through which bacteria found in the mammalian gut produce antimicrobial peptides.
The newly identified biosynthetic pathway transforms a biologically inert peptide into structurally complex antibiotics, which they call enteropeptins. Enteropeptins are a class of ribosomally synthesized peptide natural products, referred to as RiPPs.
The core structure of these products is synthesized by the ribosome, which is limited to the 20 canonical amino acids. The Mo Lab discovered and characterized new metalloenzymes capable of converting arginine, a canonical amino acid, into N-methylornithine, a noncanonical amino acid, within enteropeptin.

Brett Covington, left, postdoctoral researcher in the Mo Lab; and Professor Mohammad Seyedsayamdost.
The lab’s paper, “Biosynthesis-guided discovery reveals enteropeptins as alternative sactipeptides containing N-methylornithine,” was published last month in Nature Chemistry.
Kenzie Clark, first author on the paper and a former graduate student in the Mo Lab, explained the core discoveries.
“The way this pathway works is that the ribosome makes a precursor peptide, which is then acted on by metalloenzymes encoded in the same gene cluster,” said Clark. “The series of metalloenzymes—in this case, there were three of them—convert arginine into N-methylornithine in a step-wise fashion to generate enteropeptin.
“The interesting thing is that the peptide just on its own does not show any biological activity. But once you add these modifications, it becomes this active biomolecule that potently inhibits the growth of the producing strain.”

Kenzie Clark, former graduate student in the Seyedsayamdost lab
Brett Covington, a postdoctoral researcher in the lab and co-author on the paper, weighed in on this finding.
“That’s a trend we are seeing with a lot of these RiPP products that we’ve uncovered: they have very narrow-spectrum activity and they tend to inhibit the growth of the organism that’s making the compound,” he said. “That was the case here with enteropeptin. It really only inhibited the Enterococcus that made enteropeptin. Why these Enterococci make an antibiotic peptide to inhibit their own growth is a question that we’re pursuing. They are likely involved in generating a more recalcitrant bacterial population.”
Working Through Gene Clusters
The research fits neatly into the mission of the Mo Lab, which seeks to uncover new bacterial natural products and understand how these products are biosynthesized.
Back in 2018, the Mo Lab published a paper in the Journal of the American Chemical Society (JACS) in which they used bioinformatics to uncover 600 RiPP gene clusters from streptococci that utilize radical S-adenosylmethionine (rSAM) enzymes, one of nature’s largest-known family of enzymes. While it’s a huge group with over 500,000 members and is found in all three kingdoms of life, most of these enzymes are not well understood.
The lab grouped their 600 newly discovered RiPP gene clusters into 16 subfamilies based on similar precursor peptide sequences. They began working through these families in the lab, uncovering new reactions and interesting chemistry along the way.
“It’s a new approach to natural product discovery in that you start with the biosynthetic gene cluster and then you do in-depth analysis of the reaction of each enzyme to understand all the different transformations that are going on,” said Covington. “Then, we search the bacterial host for the mature product, which incorporates all these different enzymatic transformations. It’s a unique approach.”
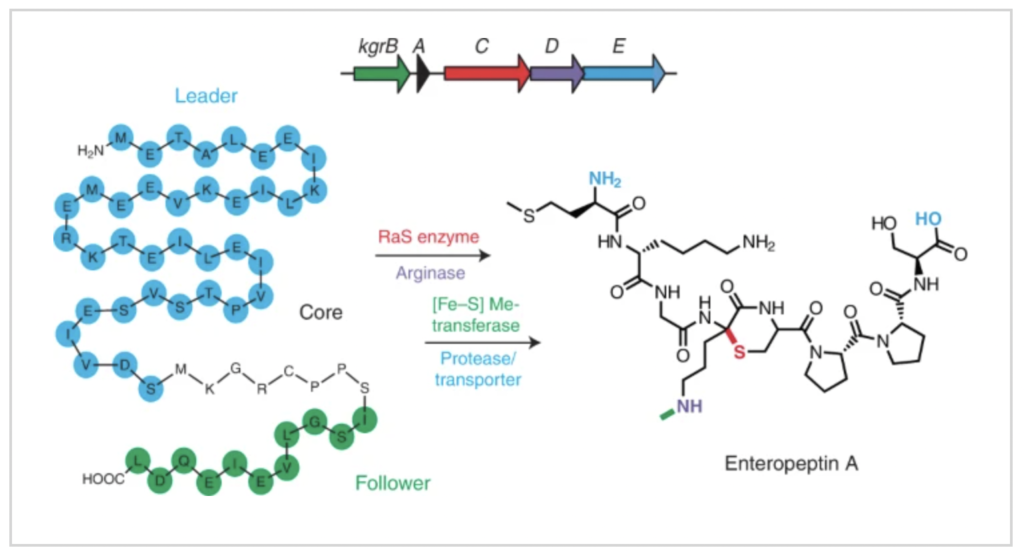
Bacterial genes kgrA-E, shown as arrows at the top, encode for the enteropeptin precursor peptide and four tailoring enzymes. These enzymes modify the core and remove the leader and follower segments of the precursor to produce enteropeptin A.
So far, they’ve only worked with clusters that code for a single metalloenzyme. This latest work targets multi-step pathways with more complex chemistries that have not just one interesting reaction, but three catalzyed by three different classes of metalloenzymes. Two are iron-dependent and one is manganese-dependent.
“One of these metalloenymes generates a very reactive intermediate that’s able to do really challenging chemical reactions, for example carbon-carbon bond formation at unactivated carbon centers,” said Clark. “A lot of the structures they generate would be very challenging for synthetic chemists to make.
“But what’s also interesting was the third enzyme in the pathway. It wasn’t annotated as a known enzyme family. It ended up using a four-iron-four-sulfur cluster and cofactor SAM to N-methylate ornithine, a novel and really interesting modification reaction.”
Covington added: “That’s one of the things I enjoy most about working here in the Mo lab, the thrill you get when you finally uncover one of these products. All the work is done on the bench to understand the reaction in the test tube, and then it’s really gratifying when you see it match up with what’s going on inside the actual bacteria.”
Journal paper here: https://www.nature.com/articles/s41557-022-01063-3
“Biosynthesis-guided discovery reveals enteropeptins as alternative sactipeptides containing N-methylornithine,” was authored by Kenzie Clark, Brett Covington, and Mohammad Seyedsayamdost. Support for this research was provided by the Edward C. Taylor-Eli Lilly Fellowship in Chemistry and the National Science Foundation (NSF CAREER Award1847932).
