Michael Hecht
Contact:
Michael Hecht
Professor of Chemistry
[email protected]
Frick Laboratory, 330
609-258-2901
Faculty Assistant:
Julia Wlodarczyk-Mingozzi
Faculty / Grants Assistant
[email protected]
(609) 258-3674
Frick Laboratory, 389
Research Focus
From Protein Design to Artificial Genomes. Scientific understanding at the interface of chemistry and biology has advanced to a point where we can consider the possibility of sustaining life using molecular parts that did not evolve in nature, but which are designed and synthesized in the laboratory. Although myriad forms of life evolved over billions of years, the biosphere has sampled only a minuscule fraction of the possible genes and proteins that could be constructed. Are these biologically selected sequences somehow special? Or is it possible to sustain life using a ‘molecular parts kit’ designed de novo?
Our research addresses these questions by going beyond the limited collection sampled by nature; beyond what exists (or existed in the past) in living systems. We explore artificial biological information (genes) and macromolecules (proteins) to probe the ability of these novel genes and proteins to perform biochemical activities, and ultimately to sustain life.
Our work toward artificial biology is enabled by methods we developed to design and construct combinatorial libraries of novel proteins that fold into stable structures. With collections of millions of well-folded de novo proteins, we explore a range of questions at the interface of chemistry and biology. For example, we can test the chemical and structural requirements for protein design while also probing the biological implications of proteome design. Moreover, because our designed proteins are expressed from synthetic genes cloned into living cells, we can begin to construct ‘artificial genomes’ comprising sequences (genes and proteins) that never existed before in biology.
Recent results show that these novel macromolecules – which bear no resemblance to natural genes or proteins – can provide some of the functions necessary to enable cell growth. These functions include enzymatically catalyzing essential biochemical reactions, as well as artificial gene regulators that rewire gene expression and cell physiology.
Our ongoing research demonstrates that the molecular ‘parts kit’ for life need not be limited to genes and proteins derived from nature; and artificial genomes capable of sustaining life may soon be within reach.
Selected Publications
Schnettler JD, Wang MS, Gantz M, Bunzel HA, Karas C, Hollfelder F, Hecht MH (2024). Nature Chemistry. Selection of a Promiscuous Minimalist cAMP Phosphodiesterase from a Library of De Novo Designed Proteins. In Press.
Kodai Kurihara K, Umezawa K, Donnelly AE, Sperling B, Liao G, Hecht MH, Arai R (2023) Crystal Structure and Activity of a De Novo Enzyme, Ferric Enterobactin Esterase Syn-F4 Proc. Natl Acad. Sci.(USA). 120 No. 38 e2218281120 https://doi.org/10.1073/pnas.2218281120
Spangler LC, Yao Y, Cheng G, Yao N, Chari SL, Scholes GD, Hecht MH (2022) A de novo protein catalyzes the synthesis of semiconductor quantum dots. Proc. Natl Acad. Sci.(USA) 119, e2204050119 https://doi.org/10.1073/pnas.2204050119
Wang MS, Hecht MH (2020) A Completely De Novo ATPase from Combinatorial Protein Design J. Am. Chem. Soc. 142, 36, 15230–15234. https://dx.doi.org/10.1021/jacs.0c02954
Wang MS, Hoegler KJ, Hecht MH (2019) Unevolved De Novo Proteins Have Innate Tendencies to Bind Transition Metals. Life 9(1), 8; doi: 10.3390/life9010008
Donnelly AE, Murphy GS, Digianantonio KM, Hecht MH (2018) A De Novo Enzyme Catalyzes a Life-Sustaining Reaction in E. coli. Nature Chemical Biology 14, 253–255 DOI: 10.1038/nchembio.2550
Hecht MH, Zarzhitsky S, Karas C, Chari S (2018) Are Natural Proteins Special? Can We Do That? Current Opinion in Structural Biology 48, 124-132. DOI : 10.1016/j.sbi.2017.11.009
Digianantonio KM, Korolev, M, Hecht MH (2017) A Non-Natural Protein Rescues Cells Deleted for a Key Enzyme in Central Metabolism. ACS Synthetic Biology 6, 694–700. DOI: 10.1021/acssynbio.6b00336
Digianantonio KM, Hecht MH (2016) A Protein Constructed De Novo Enables Cell Growth by Altering Gene Regulation Proc. Natl Acad. Sci.(USA) 113, 2400–2405. doi: 10.1073/pnas.1600566113
Hoegler KJ, Hecht MH (2016) A De Novo Protein Confers Copper Resistance in Escherichia Coli. Protein Science 25, 1249-1259. DOI: 10.1002/pro.2871
Fisher MA, McKinley KL, Bradley LH, Viola SR & Hecht MH (2011) De Novo Designed Proteins From a Library of Artificial Sequences Function in Escherichia Coli and Enable Cell Growth. PLoS ONE 6(1): e15364. doi:10.1371/journal.pone.0015364
Kamtekar S, Schiffer JM, Xiong H, Babik JM & Hecht MH (1993) Protein Design by Binary Patterning of Polar and Non-Polar Amino Acids. Science 262, 1680-1685. DOI: 10.1126/science.8259512
Related News
Summer 2024 Series: #AnnotatedChemistry One
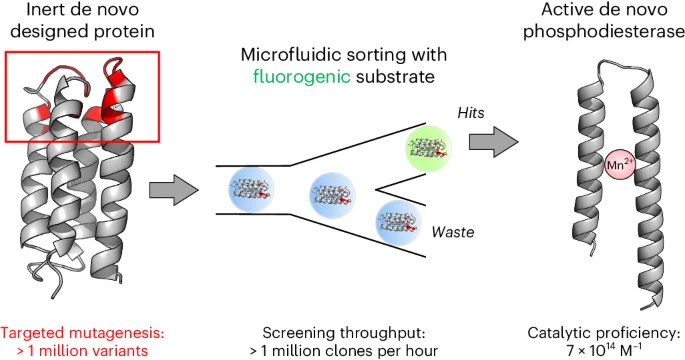
Hecht Lab finds truncated de novo proteins that act as biological catalysts