Hecht Lab finds truncated de novo proteins that act as biological catalysts
NAME OF PAPER: “Selection of a promiscuous minimalist cAMP phosphodiesterase from a library of de novo designed proteins.”
JOURNAL: Nature Chemistry, May 3, 2024. DOI: https://doi.org/10.1038/s41557-024-01490-4
AUTHORS: J. David Schnettler; Michael Wang; Maximillian Gantz; H. Adrian Bunzel; Christina Karas; Florian Hollfelder; Michael Hecht.
WHAT IT IS: In research that contributes to our understanding of how random amino acid sequences might evolve over eons into functional enzymes, the Hecht Lab reveals a truncated, structurally dynamic de novo enzyme that catalyzes an important biological reaction. The enzyme accelerates hydrolysis, a common form of chemical reaction, in the presence of manganese ions. This is significant because metals are thought to be highly important in chemistry that occurred as early life forms were emerging.
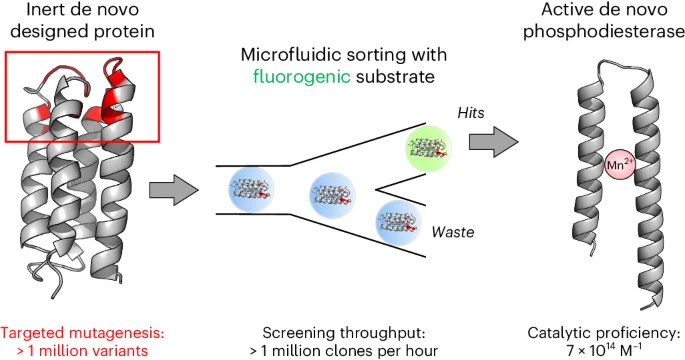
HOW RESEARCHERS DID IT: A chance conversation more than five years ago between then-graduate student David Schnettler of the Hollfelder Group at the University of Cambridge, and Michael Hecht, who was lecturing at Cambridge, led to a new application for the Hollfelder lab’s expertise with ultrahigh-throughput droplet microfluidics. Using a large library of Hecht’s de novo amino acid sequences, Schnettler’s process returned hits from a collection of 108 cells expressing novel protein sequences with an acquisition of function involving aberrant truncations in the protein sequence – as much as 40% of each protein chain was deleted.
While Hecht said he would likely have ended the investigation there—“In the normal world, a truncation is a dealbreaker”—a talented former graduate student, Michael Wang, stepped up to investigate the terminations. Wang, Schnettler, and their co-authors postulate in Nature Chemistry that these truncated sequences may resemble the kind of enzymes that nature found useful in early evolution: shorter, simpler, more versatile building blocks that had utility as primitive catalysts.
The characterization further showed that the truncated protein breaks down cyclic AMP, a signaling molecule in biology. And it does so with a high activity relative to background. The novel truncated protein was therefore named “mini-cAMPase.”

Professor of Chemistry Michael Hecht.
QUOTE FROM P.I. MICHAEL HECHT: “Think about early, early evolution, which is what we are doing in my lab. Our proteins are entirely non-natural, so we’re asking if we can come up with something that never existed in nature that can do catalysis. Our results seem to indicate that early evolution selected for activity with small building blocks that evolved into the big, complicated proteins we have today. It worked with simple, structurally dynamic enzymes that were well suited to evolution. With these truncations, you have smaller building blocks, and maybe they really are better for starting materials. They’re easier to work with. They’re more promiscuous.
“We’re taking semi-random sequences that had no activity and looking for the arising of the beginning of activity. One might think that random, arbitrary sequences wouldn’t have any activity. But at the end of the day, that’s what we got. A dynamic, truncated version that accelerates reactions a billion-fold relative to background.”

Michael Wang, a former graduate student in the Hecht Lab, now with a startup in California.
QUOTE FROM CO-AUTHOR MICHAEL WANG: “This project, which came out of a collaboration with the Hollfelder Group across the pond, revealed interesting ways in which a protein can adopt a new function. Since we can design proteins, we set out to look at how this kind of gain-of-function can emerge within a model system. The surprising discovery, in my opinion, was that the most successful variant found a new job in a very counterintuitive way: by truncating, taking away half the protein. To personify them a bit, they needed to lose a lot of the baggage holding them back.
“The kind of function we found is also exciting. We found that this very truncated sequence could perform phosphodiester hydrolysis—a reaction that is ubiquitous in life on Earth.
“As to why we dove into this despite the messiness, my rationale was, ‘where there’s smoke, there’s fire.’ We had a pretty clear data set showing that these truncations were being enriched. There were challenges. But once the first few experiments started to work, I think it became more interesting to think about why we were getting this surprising truncation.
“Thinking back, there would have been a lot less to discover if everything had gone according to plan.”
(Wang is an Enzyme Development Scientist at ZymoChem Inc., a California-based biochemical company.)
FUNDING: The work in Princeton was supported by NSF grant MCB-1947720 to M.H.H.
READ THE FULL PAPER: “Selection of a promiscuous minimalist cAMP phosphodiesterase from a library of de novo designed proteins.”
