Jacobs Lab identifies how “bridge” proteins control condensate structure
A few years ago, researchers in the Jacobs Lab went looking for an “easy” way to tune the interface between biomolecular condensates: is there some mechanism, they asked, that accounts for why these liquid-like droplets dock with each other, or don’t? And can it be manipulated?
In their new paper out last month in PRX Life, they identified how proteins can serve this very function. Just small changes in the concentration of a shared, intracellular protein can determine whether droplets dock to form larger, more complex morphologies or whether they disperse.
Using calculations that capture the essence of the physical chemistry at work, researchers were able to make theoretical predictions about condensate structure and the protein interaction network that plays a part in it.
“Why is this useful to know? Because structure is tightly linked with function,” said graduate student Tianhao Li, first author on the paper.
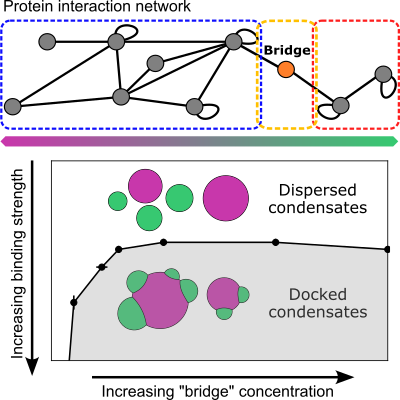
“We’re curious about what drives the formation of different morphologies, and whether we can make use of this process to do something we want. Nature does this, but we don’t understand it. So as a theorist, I want to understand this process on a fundamental level. It’s a very interesting question.”
The discovery of this mechanism adds important knowledge to our understanding of intracellular organization, particularly as condensates build into ever more complex structures.
“We have a variety of proteins that interact with each other to form this network,” said William Jacobs, assistant professor in the Department of Chemistry. “We’ve found that the one we’re calling the bridge protein resides in the middle of it. By changing the concentration of this one protein by a very small amount, you can trigger the transition from being a stable interface to being dispersed. Or vice versa.
“The key thing we’re looking for is an easy way of making this change from dispersed to stable. And by ‘easy,’ we mean something that biology could tune without a huge amount of effort. We found that all you have to do is express this bridge protein at a relatively low concentration, and it changes whether you have a dispersed or a docked morphology.”
The paper, Predicting the Morphology of Multiphase Biomolecular Condensates from Protein Interaction Networks, authored by Li and Jacobs, was published on June 20.
Satisfying a CAREER Grant
Biomolecular condensates are liquid-like structures comprised of proteins, RNA, and nucleic acids. They spontaneously assembly into forms best described as droplets within cells. These droplets have a number of biological functions and behaviors; some known, some still to be worked out.
One of the things to be worked out—and which the Jacobs Lab is particularly dedicated to finding out—is why they stick together to create “multi-phasic structures.” Or, conversely, why they disperse so that they’re not in contact but instead distributed throughout the cell.
What determines this organization?

Graduate student Tianhao Li of the Jacobs Lab, first author on the research published last month in the journal PRX Life.
“We had a hypothesis that this bridge protein might play an important role in regulating this morphology. And it makes physical sense, right?” said Li. “But based on the hypothesis, we needed to answer the question – how does this bridge protein change the morphology? We tried different models, some more complex, some less. But I think the final conclusion for our paper is that a simplified model, probably the simplest you can imagine, can probably capture this behavior pretty well.
“And that is, you just overexpress this protein and see what happens. That gives us a consistent picture that is very assuring.”
The project was part of the proposal to Jacobs’ National Science Foundation CAREER grant in 2022. The proposal, Programmable Control of Biomolecular Condensate Self-Assembly, established an early, core mission of his lab: understanding the mechanisms of biological self-assembly; and specifically, the formation of condensates and the forces driving their morphologies.
At the time, Jacobs summarized his motivation by saying: “The more we look, the more we find.” It has continually borne fruit for the lab in research papers and in collaborations with the Brangwynne Lab in Chemical and Biological Engineering.
“For this paper, we based our work on previous experimental results,” Li explained. “From there, we generated a series of predictions that might be interesting or useful for the experimentalists to test.
“So, we have those predictions. Now, they remain to be tested.”
Predicting the Morphology of Multiphase Biomolecular Condensates from Protein Interaction Networks was supported by funding from an NSF CAREER Grant.
