Kleiner Streamlines RNA-protein Interaction Analysis with TRIBE-ID

How RNA behaves in cells is largely governed by its interactions with a class of binding proteins called RBPs. There are between 2,000 and 3,000 RBPs in the human proteome, but scientists know little about them. Only a fraction has been well-characterized.
This is partly because of methodology. Right now, the main techniques used to investigate RNA-RBP interactions are labor- and time-intensive and often unreliable. More efficient methods are in development but have limited application – the shortest duration for these assays is around 16 hours, which is a great deal of time in the life of a cell.
Two years ago, the Kleiner Lab stepped into this landscape with a simple question: Is there an easier way to measure dynamic protein-RNA interactions?
This week, the lab announces the development of TRIBE-ID, a streamlined method for studying RNA-protein interactions in a cell with temporal precision through RNA editing and small-molecule induced dimerization. Inspired by the success of proximity-labeling technologies, TRIBE-ID uses a native enzyme from the cell to deposit tags that mark the RNA transcripts participating in RNA-protein binding events.
TRIBE-ID gives scientists broader control over the length of the assay, down from a day to a few minutes, which allows dynamic changes to be tracked closely.
In the first application of the methodology, TRIBE-ID was used to profile substrates of the RBPs G3BP1 and YBX1 during the formation of stress granules. Researchers discovered that the presence of stress granules has minimal impact on G3BP1’s capacity to perform one of its primary tasks: the stabilization of RNAs.
The team’s work, “Profiling dynamic RNA-protein interactions using small molecule-induced RNA editing,” appears this week in Nature Chemical Biology.
“There are established methods to capture RNA-protein interactions. In fact, many,” said Kyung “Josh” Seo, a sixth-year graduate student in the Kleiner Lab and the paper’s first author. “But often these methods require really challenging biochemical steps that can introduce unwanted variables. We always want easier alternative methods, so we turn to RNA-editing enzymatic approaches. They offer an easy readout by sequencing, but one big issue is the lack of time resolution as the fusion protein is often active for many hours.
“So, what we did was separate the protein of interest and the RNA-editing enzyme. We constructed it this way so that when we add a small molecule, they come together. And that event occurs within the timescale of minutes, which allows us to assay really fast, dynamic interactions. In our paper, we’ve shown that we can get that down to 15 minutes.”
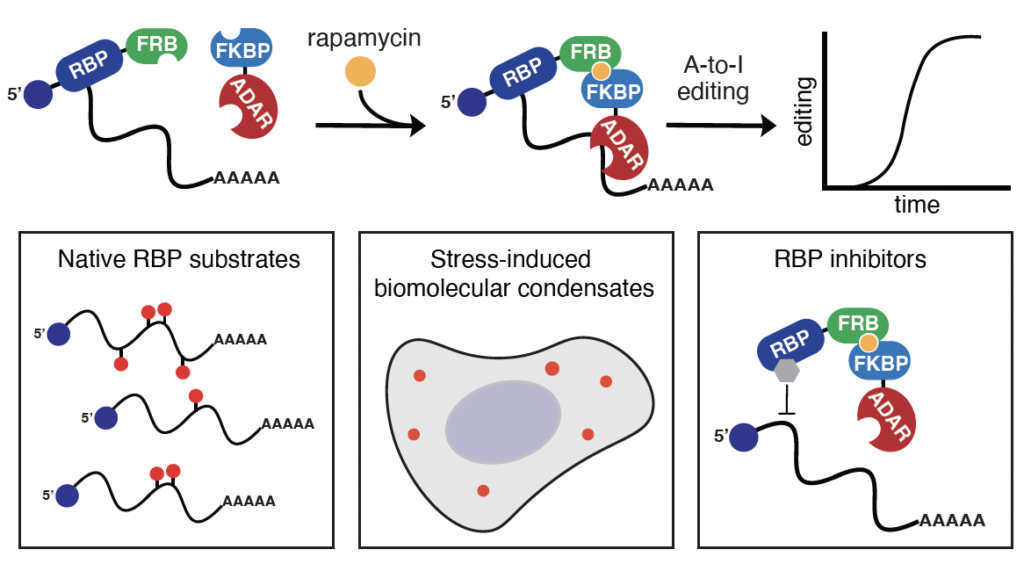
By rapamycin-mediated dimerization, TRIBE-ID captures dynamic RNA-protein interactions in cellular condensates and small molecule modulated interactions.
Assistant Professor of Chemistry Ralph Kleiner added: “The ability to use RNA editing to look at native RNA-protein interactions is going to be incredibly powerful, since it allows us to use high-throughput sequencing as the primary downstream readout to measure biochemical events occurring in the cell.
“It simplifies a lot of these analyses, which makes data more reliable across multiple experiments. Now we have the ability to do that in a temporally defined manner. That gives you the ability to look at highly dynamic processes and interactions between RNAs and proteins that change rapidly on short timescales.”
How they did it
Until recently, RNA protein-binding events were investigated primarily through a process called UV-photo crosslinking, in which the cell is irradiated to freeze all the interactions. After that, scientists have to perform some fairly elaborate work to isolate individual RNA-protein complexes and find out what those interactions are.
TRIBE-ID gets around this by marking RNA-RBP interactions in the cell dependent upon the addition of a small molecule compound called rapamycin, a kind of molecular glue.
“So imagine before we add the small molecule, you have two things floating around inside the cell: the RBP and the RNA editing enzyme that we add. They don’t ever see one another. They have no reason to be in the same place,” said Kleiner. “But once you add the small molecule, this molecular glue—also called a chemical dimerizer—brings them together very quickly.
“Now the enzyme and the RBP are in the same place. Wherever the RBP goes the enzyme follows, and what the enzyme does is start depositing these tags onto the RNA that the RBP associates with. The tags are actually just sequence mutations. This is really kind of the power of the system since we can identify these mutations easily with RNA sequencing, and we don’t have to isolate the native RNA-protein complexes.”
In fact through this process, the lab provides new insights into G3BP1.
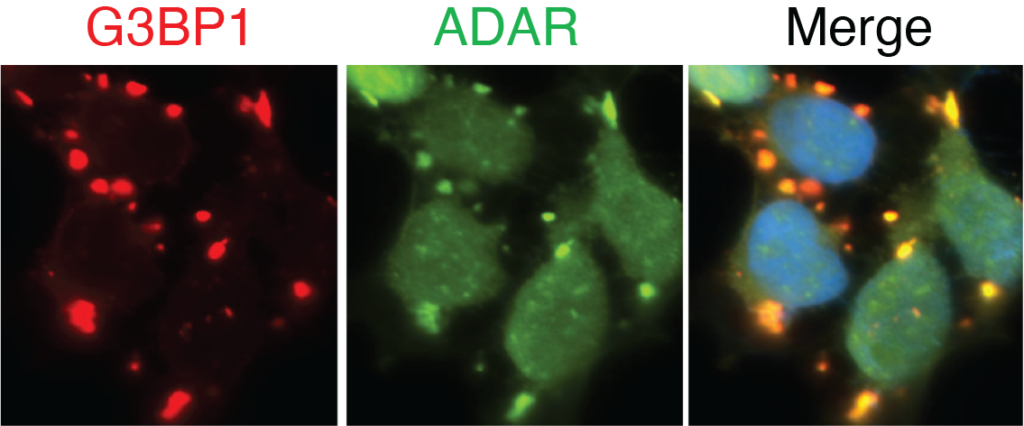
Visualization of TRIBE-ID with imaging analysis of G3BP1 and ADAR protein constructs in stress granules.
An abundant RBP, G3BP1 interacts with thousands of RNA transcripts. At the same time, it somewhat idiosyncratically forms condensates known as stress granules when cells are exposed to different types of stress.
Stress granules are micron-sized cellular structures that can be associated with certain types of neurodegenerative conditions, so scientists are intrigued by their role: are they simply artifacts that the cells carry, or do they have a greater significance? What are the biophysics behind their formation? One of the ideas around the formation of these structures is that they serve a protective role for RNAs during periods of stress.
While many questions about stress granules persist, TRIBE-ID was able to demonstrate that stress granules have little impact on the ability of G3BP1 to stabilize RNAs.
“We were able to map the substrates of G3BP1 and determine that it functions to stabilize the RNA transcripts it interacts with,” said Kleiner. “This is important for the expression of proteins encoded by these RNAs. But as it turns out, G3BP1 does this completely independently of these stress granules—they appear to have nothing to do with RNA stability.”
“So, we’ve punched a big hole in one of the leading hypotheses for what stress granules are doing. It was an important insight for us in guiding our studies further.”
This research was supported by funding from the U.S. Department of Health & Human Services, National Institutes of Health, National Institute of General Medical Sciences (grant R01 GM132189).
