Mass Action Controls Metabolite Homeostasis, Rabinowitz Finds
We tend to think of the human body as operating out of a sense-and-respond paradigm. For example, you eat sugar, the pancreas senses the intake, and it responds by releasing the hormone insulin to keep the body’s glucose levels in check.
Scientists have long wondered whether there is a similar regulatory strategy for clearing other metabolites, like amino acids or citrate, that keeps blood levels in a healthy range.
The Rabinowitz Lab reports instead that metabolite homeostasis in animals doesn’t require any sensing at all: a chemical principle known as mass action oversees homeostasis, automatically clearing many of these circulating metabolites when they show up in the bloodstream without the production of a signal hormone.

Joshua Rabinowitz, professor of chemistry and the Lewis-Sigler Institute for Integrative Genomics, and director of the Ludwig Institute for Cancer Research Princeton Branch, and P.I. on new research on how animals maintain homeostasis.
Circulating metabolites feed our organs a continuous supply of essential nutrients and building blocks. Understanding their regulation and control has important implications for the treatment of disease. In order to responsibly manipulate amino acid levels or carbocyclic acid levels to bolster the immune system or impair tumor growth, for example, we have to understand the basic rules by which these levels are governed physiologically.
The lab’s recent paper in Nature Metabolism, “Circulating metabolite homeostasis achieved through mass action,” lays out the biology in the first quantitative study in animals to demonstrate the relationship of mass action to the preservation of homeostasis.
More broadly, it illustrates how chemistry can illuminate and even solve longstanding biological questions.
Researchers on the project are from the Department of Chemistry, the Lewis-Sigler Institute for Integrative Genomics, and the Ludwig Institute for Cancer Research Princeton Branch and the Department of Molecular Biology. Xiaoxuan Li, an advisee of Rabinowitz who earned her Ph.D. in molecular biology, is lead author on the paper.
“In layman’s terms, when you eat a meal and you have an influx of amino acids from protein or acetate from vinegar, your body naturally adapts to clear the excess levels from the bloodstream. This happens simply via chemical machinery that makes the nutrients go away in proportion to how abundant they are. It doesn’t take any regulation or sensing – you never sense these things. You eat them and your body consumes them,” said Joshua Rabinowitz, professor of chemistry and Lewis-Sigler.
For fundamental aspects of metabolism, such mechanisms are apparently unnecessary, he said.
“With this research, we did not find any mechanism in the body to say: stop releasing a particular amino acid into the bloodstream because you have too much of it. There are parts of biology, parts of our bodies, that work in a simple way. This is a nice example of that,” added Rabinowitz, who is also the director of the Ludwig Princeton Branch. “Things that are simple are robust, and this is probably part of why there aren’t a lot of people suffering from amino acid imbalance.”
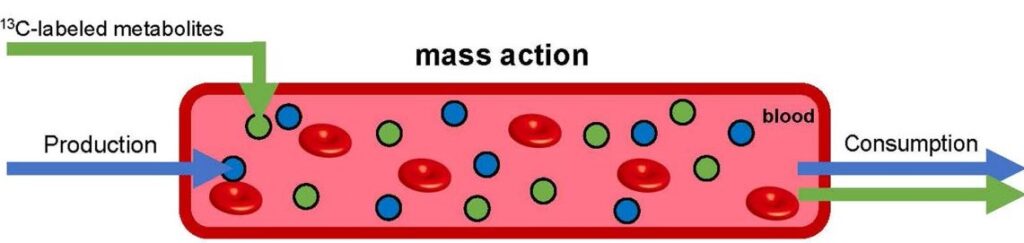
Mass action clears metabolites from the bloodstream in linear proportion to their concentration. When the concentration of a particular metabolite is augmented by infusing it (in isotope labeled form, so that it can be traced, green), production and consumption of the natural unlabeled metabolite (blue) remains unchanged. This simple mechanism for maintaining homeostasis does not require active sensing to maintain healthy metabolite levels in the blood (red).
Perturbative Isotope Labeling
The lab employed perturbative isotope-labeled metabolite infusions, increasing the overall metabolite concentrations substantially in subject mice, in order to allow the quantification of the synthesis and breakdown fluxes. Researchers delivered a so-called 13C-tracer (a heavy but not radioactive form of the nutrient) in a “perturbative” way – to agitate and spur a reaction that could be investigated through mass spectrometry.
When a metabolite concentration increases significantly, the body can counteract it through two means: by reducing the endogenous production, or by trying to burn more quickly.
“We tested this idea in abundant circulating metabolites and found that the consumption fluxes of most metabolites are directly proportional to the metabolite concentration, and for everything but glucose, the production fluxes remain at a similar level,” said Xianfeng Zeng, one of the paper’s authors and a former graduate student in the Rabinowitz Lab. “That indicates that the body maintains the homeostasis by increasing consumption, but not by reducing production.
“This is one of the most important and profound results from our lab, it explains the biology in a clean and quantitative way.”
Li emphasized the quantitative foundation of the work. “Our study really viewed this problem from a systemic perspective, not limited to a single metabolite/metabolite group, and recognized the importance of the simple but powerful rule of mass action. Biology can be complicated and hard to quantify with high variance, so we want to emphasize the quantification in this paper and show people how a simple equation can explain such complex systems like mammals.”
Institutional collaborators on the work include the Department of Molecular Metabolism at Harvard, the Department of Nutritional Sciences at Rutgers University, and the Department of Biological Chemistry at the University of California, Irvine.
Read the full paper: https://www.nature.com/articles/s42255-021-00517-1
Appearing in Nature Metabolism (doi.org/10.1038/s42255-021-00517-1),“Circulating metabolite homeostasis achieved through mass action,” was authored by Xiaoxuan Li, Sheng Hui, Emily Mirek, William Jonsson, Tracy Anthony, Won Dong Lee, Xianfeng Zeng, Cholsoon Jang, and Joshua Rabinowitz. The research was supported by NIH Pioneer (no. 1DP1DK113643) and Paul G. Allen Family Foundation grants (no. 0034665) and Ludwig Cancer Research.
