Muir Lab creates protein activation platform with “exquisite” precision
When sailors need to chart a ship’s location without using electronics, the navigational skill of triangulation—which any sailor worth their salt learns early—allows them to select three reference points whose intersecting lines can determine the exact position.
It’s a fitting analogy for the Muir Lab’s latest research, which uses two molecular reference points to zero in with “exquisite targeting” on the precise location of specific cell sites within complex mixtures.
That is significant because it’s never been done before with quite this level of precision—specificity is a key goal in biochemistry—and because their platform enables the functionalization of a new protein on the cell surface.
Now, using this platform, scientists will be able to send therapeutics with exactitude, opening up biological applications for a range of unprecedented tasks and transforming how we interrogate, manipulate, and treat specific cells.

Tom Muir, the Van Zandt Williams Jr. Class of 1965 Professor of Chemistry, in his lab.
Called SMART, for Splicing Modulated Actuation upon Recognition of Targets, the lab’s startling technology combines split inteins and simple logic gating to create a programmable system for cell-specific protein ligation that is autonomous: program it and it will do the rest.
SMART was introduced in Nature last week: Programmable protein ligation on cell surfaces.
“Our goal was to develop a modular and tunable approach that would allow us to use specific features of cell surfaces to compute, using various logic operations, the post translational synthesis of a protein output. The SMART system does just that,” said Tom Muir, the Van Zandt Williams Jr. Class of 1965 Professor of Chemistry. “The study involved a great deal of protein engineering and protein chemistry, and I am very proud of the ingenuity and resourcefulness of the team.
“The study has opened many new projects in the Muir Lab that I’m super excited about.”
Split inteins enable the platform
Christian Kofoed, first author and a former postdoc in the Muir Lab, said the platform was made possible through the lab’s caged split inteins tool, which splice and activate a new protein exactly where they’re programmed to do it.
“Most systems like this rely on external triggers, like a drug or light, which are supplemented by the user,” said Kofoed, now an assistant professor at the University of Copenhagen. “I joined the Muir Lab with a different idea: what if we could make these systems respond to their environment instead, and activate themselves only when the right context is detected? That became the foundation for SMART.”

Christian Kofoed, first author on the new Muir Lab paper and a former postdoc in the lab.
“SMART uses surface proteins on a cell as the input signal. When the right ones are present, they bring the fragments together, triggering the splicing reaction and creating a new, active protein as the output. In a way,” he added, “it’s molecular decision-making. The components do the sensing and acting themselves.”
In other words, scientists can take any protein and tie its activation to a particular cell type or condition and then simply let SMART off the leash to do its thing.
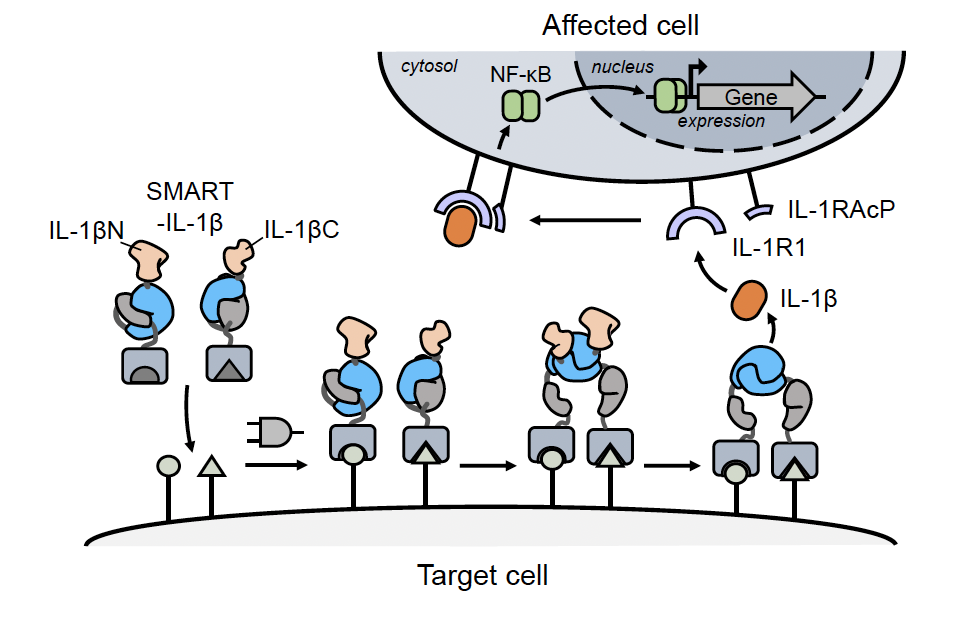
The proving ground was Interleukin-1β (IL-1β), a key protein in the human body that is produced by white blood cells and a major focus in immunotherapy.
“We wanted to show that SMART could work in a paracrine-like way, where a response in one cell triggers a change in another,” Kofoed explained. “With SMART-IL-1β, we programmed one cell to be the ligation site for the cytokine. Once produced, it binds to receptors on neighboring cells and activates them. That kind of cell-to-cell coordination is difficult to engineer but SMART makes it possible. It’s actually quite remarkable.”
SMART operates through logic gates, in which binding vectors and activation occur on the cell surface at a preprogrammed site only, dependent on certain conditions. Its homing device—the two molecular reference points—are antigens: proteins or receptors in close proximity. When these two, specific antigens are detected by SMART, they autonomously trigger the activation of split inteins. The inteins switch on and ligate or join the protein back together to form a new, functional protein able to perform a new function.

Girum Erkalo, graduate student in the Muir Lab and co-author on the Nature paper.
“Interleukin-1β can be an effective treatment for cancer, for example,” said co-author Girum Erkalo, a rising fourth-year graduate student in the Muir Lab. “But you have to use a higher dose to get it to where it has to go, and then it just goes crazy and you can get all sorts of side effects.
“What we do is split it in two, and now that protein is non-functional. Then, when they detect those two receptors on the cell surface—and only when they detect them—they turn on and ligate this thing back together into a functional protein.
“This way, if we’re making it right there in that microenvironment, we can access something that has previously been inaccessible.”
The new protein can be a delivery system or loading dock for therapeutics, or serve as a biological tool to study the cell at that particular site, or it can be a drug itself.
The lab is already pursuing a number of investigations related to the SMART platform, which could power next-generation tools in synthetic biology, cell targeting, and microenvironment micromapping.
Funding for this research was provided by the National Institutes of Health and the Ludwig Princeton Branch of Ludwig Cancer Research.
