Princeton Chem Develops Predictive Tool For Phosphine Reactivity
Continuing in the mission to expand data science tools for discovery optimization, Visiting Research Scholar Abigail Doyle, Princeton Chemistry graduate students, and collaborators introduce a workflow that allows scientists to predict phosphine ligand reactivity on the basis of one structural parameter.
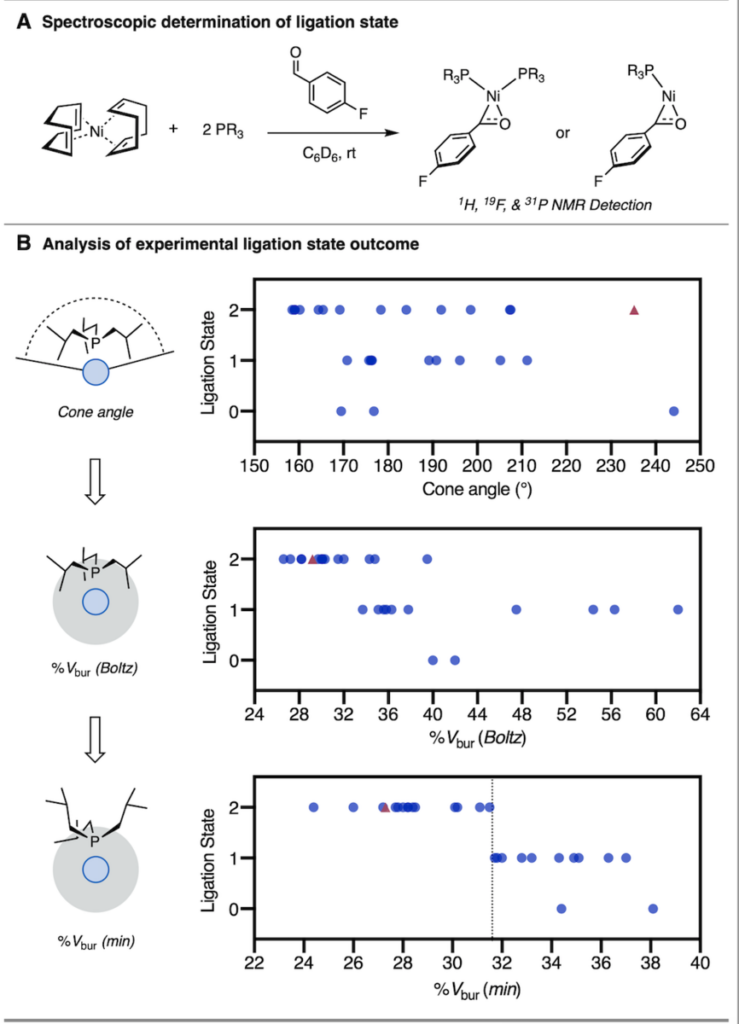
In the journal Science this week, the Doyle Lab reports that a ligand steric feature, %Vbur(min), effectively determines whether some ligands are active or inactive based on the observation of a reactivity “cliff,” or a dramatic drop off in reactivity.
The work revolves around 11 nickel and palladium-catalyzed cross-coupling datasets employing phosphine ligands. Phosphines are among the most ubiquitous ligands used in transition metal catalysis because they bind to a metal and modify its structure and reactivity.
While ligand size is known to impact these reactions, the observed reactivity cliff and corresponding steric feature, %Vbur(min), had yet to be uncovered. Most features that describe a ligand’s structure cannot define a clean divide in reactivity. But the use of %Vbur(min) allows chemists to separate active and inactive ligands computationally, before an experiment even takes place.
The research was driven by organometallic studies showing that this feature corresponds with whether or not the ligand binds 2:1 or 1:1 to the metal.

Abigail Doyle, a Princeton Chemistry visiting research scholar; and Samuel Newman-Stonebraker, a fifth-year graduate student in the lab and lead author on the paper.
“The reason this is useful is because you can compute this feature without ever having to make a ligand,” said Doyle. “So, we can draw out a structure and we can compute its %Vbur(min), and then based on this tool we can say this ligand should be active or not active.
“That can be a fairly challenging thing to do ahead of time when you’re comparing phosphines that are very diverse in their structures. It is much faster and easier to draw and compute this parameter than it would be to make the phosphine and then to evaluate it experimentally.”
The paper, “Univariate Classification of Phosphine Ligation State and Reactivity in Cross-Coupling Catalysis,” was published today in Science with research from the Doyle Lab; the Sigman Lab at the University of Utah; and Merck’s Process Research and Development group in Rahway, NJ.
The Doyle Lab moved to the Department of Chemistry and Biochemistry at the University of California, Los Angeles, this summer. Several of the lab’s graduate students, including fourth-year Julia Borowski, co-author on the research, remain at Frick Laboratory to finish out their doctoral degrees.
This latest research arose out of work done several years ago by a former graduate student who discovered several new ligands that worked well for the group’s investigations into nickel catalysis. But why were they so effective?

Julia Borowski, a fourth-year graduate student in the Doyle Lab and paper co-author, in the lab.
To find out, Borowski and Princeton Chemistry fifth-year graduate student Samuel Newman-Stonebraker tapped into a virtual library of phosphine ligand parameters, or features, built by the Sigman Lab at Utah that helps elucidate how structure relates to reactivity. They used high throughput experimentation to delve more deeply into that relationship.
“The big finding is that this discontinuity, this reactivity cliff, is on the basis of the minimum percent buried volume, %Vbur(min), of the ligands,” said Borowski. “That is a computationally derived feature of a phosphine ligand that tells you something about how much steric bulk the ligand has near the metal that it’s binding to. So, a ligand that has a high %Vbur(min) will have a lot of steric bulk around the metal and it will make it harder to attach two of them.
The %Vbur (min) descriptor is uniquely capable of predicting spectroscopic ligation state outcome, revealing reactivity cliffs in organometallic chemistry.
“And what we find is that there’s a strict cutoff where only ligands below a given value of this parameter can bind two ligands. Phosphines that have values of this parameter above that value can only bind one. It was very striking to us when we found that it wasn’t a linear trend.”
Newman-Stonebraker said there are likely two ways that chemists will want to make use of the new workflow.
“For chemists who use data-driven modeling to facilitate reaction development and optimization, the ability to organize large amounts of data into ‘bins’ based on distinct mechanistic outcomes can allow for the subsequent models to be simpler and more informative,” he said. “And for chemists interested in mechanistic organometallic chemistry, the workflow can help to uncover reactivity patterns hidden in the data, providing a roadmap for targeted study of ligand structure-reactivity relationships.”
Read the full paper in Science.
The paper was authored by Samuel Newman-Stonebraker, Sleight Smith, Julia Borowski, Ellyn Peters, Tobias Gensch, Heather Johnson, Matthew Sigman, and Abigail Doyle. Funding for the research was provided by the National Science Foundation’s Center for Computer Assisted Synthesis (CHE-1925607).
