Study reveals mechanism behind enzyme that tags unneeded DNA
Researchers have discovered the two-step process that activates an essential human enzyme, called Suv39h1, which is responsible for organizing large portions of the DNA found in every living cell.
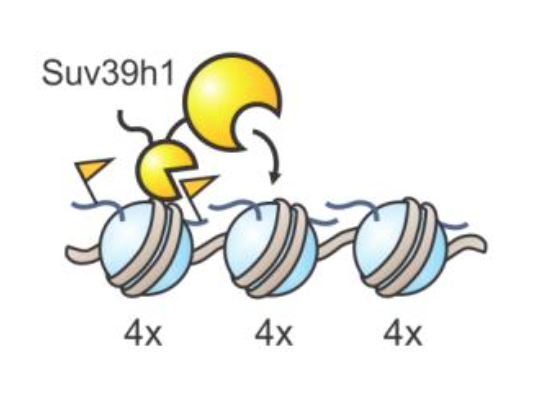
Image courtesy of the Muir lab
For any particular cell, such as a skin or brain cell, much of this genetic information is extraneous and must be packed away to allow sufficient space and resources for more important genes. Failure to properly pack DNA jeopardizes the stability of chromosomes and can result in severe diseases. Suv39h1 is one of the main enzymes that chemically mark the irrelevant regions of DNA to be compacted by cellular machinery, but little is known about how it installs its tag.
Now, scientists at Princeton have used ‘designer chromatin’ templates – highly customized replicas of cellular DNA and histone proteins, the scaffolding proteins around which DNA is wrapped – to reveal new details about Suv39h1’s mechanism. The researchers investigated how Suv39h1 employs a positive feedback loop to chemically tag thousands of adjacent histones, thus signaling the cell to stow away these underlying, unnecessary DNA sequences. The work was published in in the journal Nature Chemical Biology.
“One of the things that has always fascinated me about feedback loops is that they’re super dangerous. If you make a mistake once, you end up getting reinforcement through the feedback loop,” said Manuel Müller, a postdoctoral researcher in the Muir lab and lead author on the study. “So how does Suv39h1 keep itself in check?”
Suv39h1 had been known to possess two distinct parts, but the new research revealed how they work together in order to ‘switch on’ the enzyme. One part of the enzyme, known as the chromodomain, is constantly exposed and seeks out specific chemical tags, known as methyl groups, located at predetermined sites on histones. When the chromodomain finds these groups in the genome, it locks onto the spot and allows the other part, the enzymatic core, to install more methyl tags at adjacent histones.
“The second, anchoring step wasn’t really known before. It provides an extra level of control and allows the process to be extremely fine-tuned,” Müller said. A similar mechanism may be employed by many other enzymes operating on chromatin, given that they contain similar components of a feedback loop.
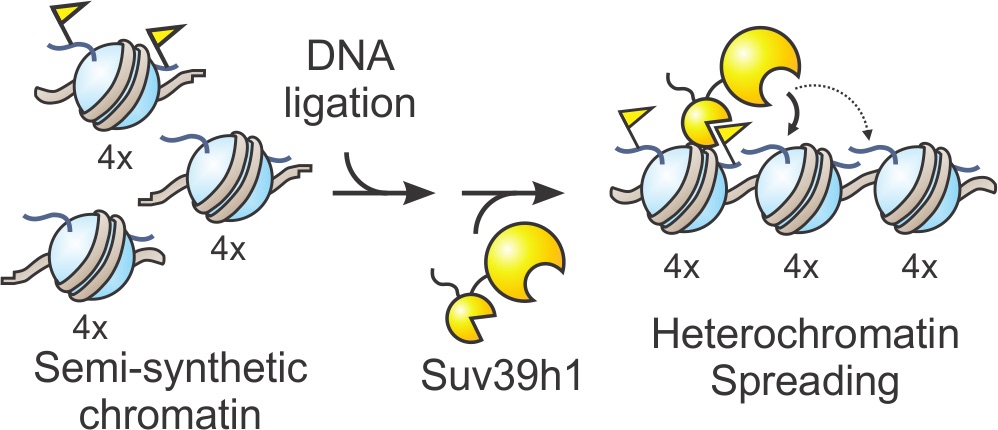
Image courtesy of the Muir lab
To understand how the enzyme carries out this process, the researchers synthesized complex chromatin templates that were three times larger than previously reported models. They divided the template into three blocks that could each be manipulated in various ways. For example, a block could be prepared with the chemical tag present, absent or mutated such that tagging can’t occur. “The different blocks should signal to the enzyme either start here or feel free to spread here or absolutely stop here,” said Glen Liszczak, a co-author and postdoctoral researcher in the Muir lab.
By rearranging the various domains, the research team observed where the enzyme spread its mark across the genome. They found that Suv39h1 preferred to spread across small distances, but that it could reach sequences further along if chromatin folding decreased the physical distance in space.
“We’ve learned something new about this enzyme, something that we couldn’t have without the pinpoint precision that the designer chromatin offers,” Liszczak said. “There are a lot of questions that our lab has been interested in that we can now start to answer.”
Read the full article here:
Müller, M. M.; Fierz, B.; Bittova, L.; Liszczak, G.; Muir, T. W. “A two-state activation mechanism controls the histone methyltransferase Suv39h1.” Nature Chem. Bio. Available online January 25, 2016.
This work was funded by the Swiss National Science Foundation (postdoctoral fellowships) and the US National Institutes of Health (R01-GM107047).
