The MacMillan Lab’s Cellular Mapping Tool Illuminates the Microenvironment
When corporate partners in the Princeton Catalysis Initiative sat down two years ago with David MacMillan, they presented him with a biological challenge at the heart of potential cancer medicines and other therapeutics: which proteins on a cell’s surface touch each other?
What they wanted was analogous to a searchlight blazing into a dark cave — something to shine a metaphorical light on a protein and its closest neighbors on the cell membrane. Large, complex molecules, proteins are the stuff of life, the very fulcrum on which everything about us turns — the way we think, the way we grow, the diseases we get. Proteins are able to determine this by sending messages to their neighbors. But while scientists could previously tell who was inside the cave, they could not tell who was standing next to whom, and thus were lacking important knowledge about these essential protein-to-protein communications.
The MacMillan Group announced in the current issue of Science that they have developed that searchlight.

The Department of Chemistry’s MacMillan Group developed a new technology called µMap that identifies proteins’ nearest neighbors on cell surfaces. The team includes (from left) Jacob Geri, Gabby Lovett, Tao Wang, Benito Buksh, Ciaran Seath, Beryl Li, Aaron Trowbridge, James Oakley, Dan Kim and David MacMillan.
The breakthrough technology, named µMap by the team of Princeton researchers and Merck scientists, uses a photocatalyst — a molecule that, when activated by light, spurs a chemical reaction — to identify spatial relationships on cell surfaces. The catalyst generates a marker that tags proteins and their molecular neighbors, which in turn enables the precise mapping of their micro-environment.
The technology could impact proteomics, genomics and neuroscience, to name a few of the more obvious fields. But the applications for fundamental biology are so wide-ranging that MacMillan, who is Princeton’s James S. McDonnell Distinguished University Professor of Chemistry, is hungry to get the technology “into everyone’s hands” to see what scientists in other fields can come up with.
“For the technologies we have right now, the problem is not whether you can tag things,” he said. “The problem is that you can tag thousands of things and so you can’t tell what’s way over there and what’s right next door. That turns out to be really, really important because molecules or proteins or enzymes that signal each other are usually right next door to each other. Well, the state-of-the-art doesn’t tell you what’s close.”
So they came up with a radical new approach.
“We did some critical experiments and immediately we could show that we were labeling things within a really short distance,” MacMillan said. “We now know exactly what’s in the neighborhood. And that’s never been done before. For biology, it’s going to be like turning on the light switch and suddenly seeing everything.”
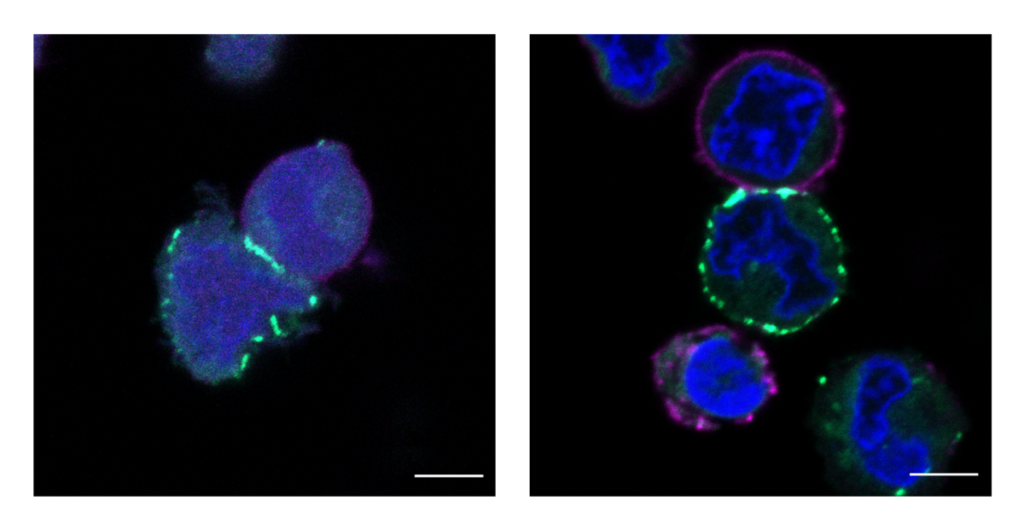
The MacMillan Group’s new technology in action: these images show cells being labeled, or highlighted in fluorescent green, by the µMap catalyst.
Merck Exploratory Science Center (MESC) scientists Rob Oslund and Olugbeminiyi Fadeyi, paper co-authors who are based in Cambridge, Massachusetts, said the technology could inspire vast new developments in biology. “Given the important role of understanding protein interactions within cellular micro-environments,” Oslund said, “this technology has the potential to be a game-changing tool for both academic and industry life science labs all over the world.”
The µMap, pronounced micro-map, identifies neighbors within a radius of 1 to 10 nanometers around a particular protein. (For reference, a human hair is about 100,000 nanometers across.) Resolution on this level identifies the 10 or 15 closest molecules.
Jacob Geri, a postdoctoral researcher in the Merck Center for Catalysis at Princeton University and a co-first-author on the Science paper with graduate student James Oakley and MESC scientist Tamara Reyes-Robles, said µMap does this by using blue light to power a catalytic reaction.
Here’s how it works: The catalyst — in this case, an organic metal compound — is selectively attached to any one of some 40,000 proteins on a cell’s surface, where it acts as a kind of antenna. Blue light, which has a very high photonic energy, serves as the trigger. When shone on the cell, this blue light is picked up by the antenna, which converts its photonic energy into chemical energy. That latent energy doesn’t cool; it doesn’t diffuse; it doesn’t wander aimlessly along the cell membrane painting everything it comes across. It just sits.
Based on a paper published some 40 years ago, MacMillan’s group came up with the idea of employing the use of an organic molecule called a diazirine that is particularly receptive to this latent energy. When a diazirine moves very close to the catalyst — within 0.1 nanometers — the chemical energy transfers to the diazirine. The diazirine in turn reacts so violently that it releases a byproduct and becomes what’s referred to as a carbene, an “angry” species that attaches itself to neighboring proteins.
“The catalyst transfers so much energy that the molecule rips itself apart to expose an incredibly unstable carbon atom, which will then just stick to anything it can,” explained Geri.
The catalyst can perform this chemical reaction many times, so the process repeats itself for all localized molecules, proteins and enzymes. Because carbenes are so short-lived — just a couple of nanoseconds — their reaction provides for a vivid, real-time snapshot of all contiguous molecules. Subsequently, researchers can quilt together a precise map of the micro-environment — the very technology scientists were looking for.
“A lot of the mechanism of disease takes place through how these cells talk to each other, and they can only talk if they’re touching,” said Geri. “That’s why the surface of the cell is so important. If they touch, they can communicate.”
He added: “We can now figure out what’s making that communication happen or what’s making that communication change. It’s really been an amazing experience, working on this.”
MacMillan’s group chose two categories of human cells to investigate. One was a class of proteins that had known interactions, selected as a kind of control group to prove that their interactions could be captured by µMap. The second group was “more interesting,” said Geri. It centered on proteins called PD-L1 and PD-1, which are associated with the body’s immune system and its response to cancer cells.
Normally, sick cells like cancer cells would present as molecular interlopers that need to be cleared by the immune system. But cancer cells are deceptive, said MacMillan. They send out a “don’t kill me” signal through a cloaking mechanism involving the PD-L1 and PD-1 axis. Since cancer therapies are successful partly based on their capacity to block that signal, scientists want to know more about how it is transmitted. Mapping the precise neighborhood is an essential early step. When researchers put the µMap catalyst on PD-L1 and PD-1, the molecules in their micro-environment are tagged. Protein-protein interactions that had previously been hypothesized could now be directly observed. And several correlations were detected that had never been conceived of.
“Now, we don’t do the cancer biology,” said MacMillan. “But we’ve invented this tool that can give you a lot of information about these cancer cells. We think that by using this information, you can start to target those proteins as a way to also remove interfering signals. And if you can remove those signals, you make your immune system better at going after these cancer cells.”
Soon after MacMillan arrived at Princeton, he began driving research in harnessing blue LED light to perform previously impossible feats of chemistry. Merck became involved in 2006, with a seed donation toward MacMillan’s research. The company has since donated additional monies, and in 2019 they announced a 10-year funding commitment towards the Princeton Catalysis Initiative, which fosters interdisciplinary collaborations to accelerate the discovery of new research areas.

Merck scientists Olugbeminiyi Fadeyi (left) and Rob Oslund of the Merck Exploratory Science Center in Cambridge, MA, collaborators.
“Our collaboration created a novel cellular chemistry approach leveraging photoredox catalysis to activate diazirines, an important class of organic molecules, in a temporal-controlled manner,” said Merck’s Fadeyi. “Because of the routine use of diazirines within chemical biology and biology, this method will be in high demand not only for protein labeling, but for identifying the binding targets of other biomolecules to elucidate their functional roles.”
He added: “The collaboration was successful due to the close interactions between Merck scientists and Dave’s lab.”
MacMillan likewise lauded the discovery as proof of the value of collaboration across academic and industry lines, like those envisioned when the Princeton Catalysis Initiative first came into being in 2018.
“As chemists, we don’t know any good questions in biology — zero,” he said. “So, you’re taking these people who know everything about biology, and they have this problem that they’re trying to solve. And it’s ultimately a great problem for a chemistry group. At the same time, it’s not a problem that a chemistry group would ever think about because they don’t know biology. You have these two different areas and you put them together and you start to realize there all these great things you can do.
“This is what I love about the social science of science,” he added. “It is absolutely a beautiful example of how it took a village to solve a problem.”
The paper, “Microenvironment mapping by means of Dexter energy transfer on immune cells,” by Jacob B. Geri, James V. Oakley, Tamara Reyes-Robles, Tao Wang, Stefan J. McCarver, Cory H. White, Frances P. Rodriguez-Rivera, Dann L. Parker Jr., Erik C. Hett, Olugbeminiyi O. Fadeyi, Rob C. Oslund and David W. C. MacMillan, appears in the March 6 issue of Science. The research was supported by Merck & Co., Inc, the National Institutes of Health (NIH) National Institute of General Medical Sciences, and an NIH postdoctoral fellowship for J.B. Geri.
